UCSC Makes Major Breakthrough with First Complete Gapless Sequence of a Human Genome
SANTA CRUZ, CALIFORNIA—UCSC Newscenter. March 31, 2022. By Tim Stephens
Karen Miga, assistant professor of biomolecular engineering at UC Santa Cruz, co-led the Telomere-to-Telomere (T2T) Consortium, which has released the first complete, gapless assembly of a human genome sequence. (Photo by Carolyn Lagattuta)
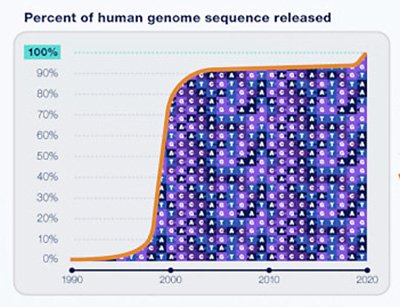
It took almost twice as long to finish the last 8% of the human genome as it did to sequence the first 92%. New laboratory and computational technologies finally enabled Miga and her colleagues to overcome obstacles such as highly repetitive DNA sequences in order to fill in the remaining gaps. (Image credit: NHGRI)
The first truly complete sequence of a human genome, covering each chromosome from end to end with no gaps and unprecedented accuracy, is now accessible through the UCSC Genome Browser and is described in six papers published March 31 in Science.
Since the first working draft of a human genome sequence was assembled at UC Santa Cruz in 2000, genomics research has led to enormous advances in our understanding of human biology and disease. Nevertheless, crucial regions accounting for some 8% of the human genome have remained hidden from scientists for over 20 years due to the limitations of DNA sequencing technologies.
In 2019, Karen Miga, assistant professor of biomolecular engineering at UC Santa Cruz, and Adam Phillippy at the National Human Genome Research Institute (NHGRI) organized an international team of scientists—the Telomere-to-Telomere (T2T) Consortium—to fill in the missing pieces. Their efforts have now paid off.
The new reference genome, called T2T-CHM13, adds nearly 200 million base pairs of novel DNA sequences, including 99 genes likely to code for proteins and nearly 2,000 candidate genes that need further study. It also corrects thousands of structural errors in the current reference sequence.
It took almost twice as long to finish the last 8% of the human genome as it did to sequence the first 92%. New laboratory and computational technologies finally enabled Miga and her colleagues to overcome obstacles such as highly repetitive DNA sequences in order to fill in the remaining gaps. (Image credit: NHGRI)
The gaps now filled by the new sequence include the entire short arms of five human chromosomes and cover some of the most complex regions of the genome. These include highly repetitive DNA sequences found in and around important chromosomal structures such as the telomeres at the ends of chromosomes and the centromeres that coordinate the separation of replicated chromosomes during cell division. The new sequence also reveals previously undetected segmental duplications, long stretches of DNA that are duplicated in the genome and are known to play important roles in evolution and disease.
“These parts of the human genome that we haven’t been able to study for 20-plus years are important to our understanding of how the genome works, genetic diseases, and human diversity and evolution,” Miga said.
Many of the newly revealed regions have important functions in the genome even if they do not include active genes.
Top of the mountain
“There is a profound advantage to seeing the whole genome as a complete system. It puts us in a position to unravel how that system works,” said David Haussler, director of the UC Santa Cruz Genomics Institute. “We’ve gotten an enormous understanding of human biology and disease from having roughly 90 percent of the human genome, but there were many important aspects that lay hidden, out of view of science, because we did not have the technology to read those portions of the genome. Now we can stand at the top of the mountain and see all of the landscape below and get a complete picture of our human genetic heritage.”
The T2T genome sequence, representing the finished CHM13 genome plus the recently finished T2T Y chromosome (CHM13 includes an X but not a Y chromosome), is now a new reference genome in the UCSC Genome Browser. The T2T sequence is fully annotated in the browser, providing an efficient way for scientists to access and visualize a wealth of information associated with genes and other elements of the genome.
“We wanted to put the information out in a way that is accessible and familiar to researchers so they can begin to build on it and use all the tools and resources the browser provides,” Miga explained.
The new T2T reference genome will complement the standard human reference genome, known as Genome Reference Consortium build 38 (GRCh38), which had its origins in the publicly funded Human Genome Project and has been continually updated since the first draft in 2000.
“We’re adding a second complete genome, and then there will be more,” explained Haussler. “The next phase is to think about the reference for humanity’s genome as not being a single genome sequence. This is a profound transition, the harbinger of a new era in which we will eventually capture human diversity in an unbiased way.”
Human pangenome reference
The T2T Consortium has now joined with the Human Pangenome Reference Consortium, which aims to create a new “human pangenome reference” based on the complete genome sequences of 350 individuals.
“Pangenomics is about capturing the diversity of the human population, and it’s also about ensuring we’ve captured the whole genome properly,” said Benedict Paten, associate professor of biomolecular engineering at UCSC’s Baskin School of Engineering, a coauthor of the T2T papers, and a leader of the pangenomics effort. “Without having a map of these difficult-to-sequence regions of the genome across multiple individuals, then we’re missing a huge amount of the variation present in our population. T2T sets us up to look across hundreds of genomes from telomere to telomere. It’s going to be great!”
The standard reference genome (GRCh38) does not represent any one individual but was assembled from multiple donors. Merging them into one linear sequence created artificial structures in the sequence. The Human Pangenome Project will make it possible to compare newly sequenced genomes to multiple complete genomes representing a range of human ancestries.
Genetic variants
An important outcome of the new T2T sequence is enabling more accurate assessments of genetic variants. When human genomes are sequenced for clinical studies to understand the role of genetic variants in disease or to study genetic diversity within and between human populations, they are nearly always analyzed by aligning the sequencing results with the reference genome for comparison. The T2T variant team documented major improvements in identifying and interpreting genetic variants using the new T2T sequence compared to the standard human reference genome.
“The new human genome is incredibly accurate at the base level, allowing us to flag hundreds of thousands of variants that had been misinterpreted by mapping them to the standard reference. Many of these new variants are in genes known to contribute to disease. We can now spot those because we have a more complete and accurate reference genome,” Miga said.
Miga’s research has focused on satellite DNA, the long stretches of repetitive DNA sequences found mostly in and around telomeres and centromeres. The centromeres separate each chromosome into a short arm and a long arm and hold duplicated chromosomes together prior to cell division.
"The centromeres play a critical role in how chromosomes segregate properly during cell division, and we've known for some time now that they are misregulated in all kinds of human diseases. But we've never been able to study them at the sequence level," Miga said. “By far the largest portion of new sequences added to the reference are centromere satellite DNAs. For the first time, we can study ‘base-by-base’ the sequences that define the centromere and can start to understand how it works.”
“Long-read” DNA sequencing technologies, such as the nanopore sequencing pioneered at UC Santa Cruz, were essential tools for the T2T Consortium. Two long-read sequencing datasets—high fidelity reads (HiFi data from PacBio systems) and extremely long reads that routinely reach lengths greater than 100,000 base pairs (ultra-long data from Oxford Nanopore devices)—enabled T2T researchers to span repetitive regions and develop strategies to ensure that the assembly was highly accurate. Miten Jain and other UCSC Genomics Institute researchers helped establish the ultra-long read protocol.
Leaders in genomics
UC Santa Cruz has a long history of leadership in genomics, starting with a seminal meeting in 1985 to discuss the sequencing of the human genome organized at UCSC by then-Chancellor Robert Sinsheimer. Haussler was invited to join the public Human Genome Project in 1999, and his team played a crucial role in its completion. At the time, James Kent, now a research scientist at the Genomics Institute and director of the UCSC Genome Browser project, was a UCSC graduate student. He wrote the code that assembled the first working draft of the human genome from data obtained by the International Human Genome Sequencing Consortium, and UCSC posted the draft online for the whole world to access. Kent then created the UCSC Genome Browser, still the most widely used platform to access the human genome.
The UC Santa Cruz Genomics Institute has continued to be at the forefront of genomics research and plays a leading role in the T2T and pangenomics efforts.
“The T2T work reflects the sustained and dedicated efforts of many people at UC Santa Cruz and elsewhere. Karen Miga has been working hard to get real centromere sequences into the human genome assemblies for a decade, and this has finally come to fruition!” said Kent. “I'm very excited to see this work combined with efforts to get telomere-to-telomere sequences from other human ancestries. We are moving quickly towards a truly complete representation of the human genome.”
Miga is a co-corresponding author of the main Science paper, “The complete sequence of a human genome,” along with Adam Phillippy at NHGRI and Evan Eichler at the University of Washington. She is also a co-corresponding author of the papers on “Complete genomic and epigenetic maps of human centromeres” and “Epigenetic patterns in a complete human genome,” and a coauthor of the papers on “Segmental duplications and their variation in a complete human genome,” “A complete reference genome improves analysis of human genetic variation,” and “From telomere to telomere: the transcriptional and epigenetic state of human repeat elements.”
Other researchers at the UC Santa Cruz Genomics Institute who are coauthors of the papers include Benedict Paten, Mark Diekhans, Erik Garrison (now at University of Tennessee Health Science Center), Marina Haukness, Miten Jain, and Kishwar Shafin. This work was supported by the National Institutes of Health.